mean returns the arithmetic mean of an array of data. >>> mean(x) x x.median returns the median of an array of data. >>> median(x) x x.variance calculates the variance of a one-dimensional array of data. >>> variance(x, mode = "Unbiased") x mode mode equal to Unbiased (which is the default value), the function variance returns the unbiased estimate of the variance. For mode equal to ML, the function returns the maximum-likelihood estimate of the variance, which is a biased estimate.
x.covariance calculates the covariance matrix of an array of data. >>> covariance(x, y = None, mode = "Unbiased") x x is a one-dimensional array and y==None, then this function returns the variance of x;x and y are one-dimensional arrays with the same length, covariance returns the covariance between x and y;x is a two-dimensional array, then covariance returns the covariance matrix of x; y is ignored.y x, or None;
modemode equal to Unbiased (which is the default value), the function covariance returns the unbiased estimate of the covariance. For mode equal to ML, the function returns the maximum-likelihood estimate of the covariance, which is a biased estimate.
x is one-dimensional and y==None: the variance in x;x and y are both one-dimensional and have the same length: the covariance between x and y;x is two-dimensional: the covariance matrix between the columns of x. Element [i,j] of the covariance matrix contains the covariance between columns x[:,i] and x[:,j].correlation calculates the correlation matrix of an array of data. >>> correlation(x, y = None, method = "Pearson") x x is a one-dimensional array and y==None, then this function returns 1.0;x and y are one-dimensional arrays with the same length, correlation returns the correlation between x and y;x is a two-dimensional array, then correlation returns the correlation matrix of x; y is ignored.y x, or None;
method"Pearson": The Pearson correlation (default);"Spearman": The Spearman rank correlation;"Intraclass": The intraclass correlation.x is one-dimensional and y==None: 1.0;x and y are both one-dimensional and have the same length: the correlation between x and y;x is two-dimensional: the correlation matrix between the columns of x. Element [i,j] of the correlation matrix contains the correlation between columns x[:,i] and x[:,j].regression returns the intercept and slope of a linear regression line fit to two arrays x and y. >>> a, b = regression(x,y) x y x and y should be equal.
a b Suppose we have a set of observations , and we want to find the probability density function of the distribution from which these data were drawn. In parametric density estimations, we choose some distribution (such as the normal distribution or the extreme value distribution) and estimate the values of the parameters appearing in these functions from the observed data. However, often the functional form of the true density function is not known. In this case, the probability density function can be estimated non-parametrically by using a kernel density estimation.
| Mnemonic | Kernel name | Function | Optimal bandwidth |
'u' | Uniform | ||
't' | Triangle | ||
'e' | Epanechnikov | ||
'b' | Biweight/quartic | ||
'3' | Triweight | ||
'c' | Cosine | ||
'g' | Gaussian |
pdf estimates the probability density function from the observed data. You can either specify the values of x at which you want to estimate the value y of the probability density function explicitly: >>> y = pdf(data, x, weight = None, h = None, kernel = 'Epanechnikov') x for you: >>> y, x = pdf(data, weight = None, h = None, kernel = 'Epanechnikov', n = 100) x contains n equidistant data points covering the domain where
is nonzero.
data weight weight==None, then each data point receives an equal weight 1.
x x, the function pdf will create x as a 1D array of n values for you and return it together with the estimated probability density function;
h h is not specified (and also if the user specifies a zero or negative h), the optimal bandwidth is used (which can be calculated explicitly by the function bandwidth);
kernel 'E' or 'Epanechnikov''U' or 'Uniform''T' or 'Triangle''G' or 'Gaussian''B' or 'Biweight''3' or 'Triweight''C' or 'Cosine'n x explicitly; passing both x and n raises an error. Default value of n is 100.
x explicitly: The estimated probability density, estimated at at the values in x;x explicitly: The estimated probability density, as well as the corresponding values of x.cpdf estimates the cumulative probability density function from the observed data. You can either specify the values of x at which you want to estimate the value y of the cumulative probability density function explicitly: >>> y = cpdf(data, x, h = None, kernel = 'Epanechnikov') x for you: >>> y, x = cpdf(data, h = None, kernel = 'Epanechnikov', n = 100) x contains n equidistant data points covering the domain where
is nonzero; the estimated cumulative probability density is constant (either 0 or 1) outside of this domain.
data x x, the function cpdf will create x as a 1D array of n values for you and return it together with the estimated cumulative probability density function.
h h is not specified (and also if the user specifies a zero or negative h), the optimal bandwidth is used (which can be calculated explicitly by the function bandwidth).
kernel 'E' or 'Epanechnikov''U' or 'Uniform''T' or 'Triangle''G' or 'Gaussian''B' or 'Biweight''3' or 'Triweight''C' or 'Cosine'n x explicitly; passing both x and n raises an error. Default value of n is 100.
x explicitly: The estimated cumulative probability density, estimated at at the values in x;x explicitly: The estimated cumulative probability density, as well as the corresponding values of x.cpdfc estimates the complement of the cumulative probability density function from the observed data. You can either specify the values of x at which you want to estimate the value y of the complement of the cumulative probability density function explicitly: >>> y = cpdfc(data, x, h = None, kernel = 'Epanechnikov') x for you: >>> y, x = cpdfc(data, h = None, kernel = 'Epanechnikov', n = 100) x contains n equidistant data points covering the domain where
is nonzero; the estimated complement of the cumulative probability density is constant (either 0 or 1) outside of this domain.
data x x, the function cpdfc will create x as a 1D array of n values for you and return it together with the estimated complement of the cumulative probability density function.
h h is not specified (and also if the user specifies a zero or negative h), the optimal bandwidth is used (which can be calculated explicitly by the function bandwidth).
kernel 'E' or 'Epanechnikov''U' or 'Uniform''T' or 'Triangle''G' or 'Gaussian''B' or 'Biweight''3' or 'Triweight''C' or 'Cosine'n x explicitly; passing both x and n raises an error. Default value of n is 100.
x explicitly: The estimated cumulative probability density, estimated at at the values in x;x explicitly: The estimated cumulative probability density, as well as the corresponding values of x.bandwidth calculates the optimal bandwidth from the observed data for a given kernel: >>> h = bandwidth(data, weight=None, kernel='Epanechnikov')
data weight weight==None, then each data point receives an equal weight 1.
kernel 'E' or 'Epanechnikov''U' or 'Uniform''T' or 'Triangle''G' or 'Gaussian''B' or 'Biweight''3' or 'Triweight''C' or 'Cosine'bandwidth returns the optimal bandwidth for the given data, using the specified kernel. This bandwidth can subsequently be used when estimating the (cumulative) probability density with pdf, cpdf, or cpdfc.
random module to draw 100 random numbers from a standard normal distribution. >>> from numpy.random import standard_normal >>> data = standard_normal(100) >>> import statistics >>> y, x = statistics.pdf(data) y as a function of x is drawn below (figure created by Pygist). 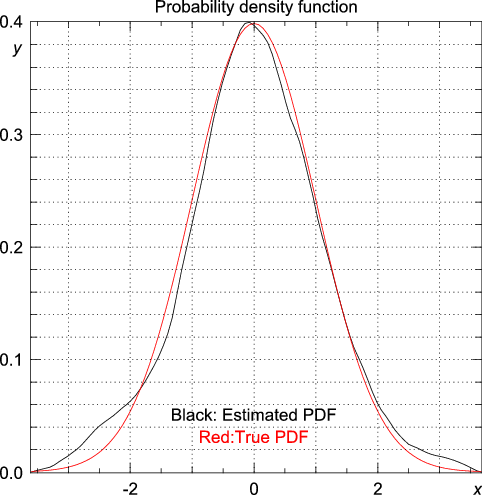
>>> y, x = statistics.pdf(data) 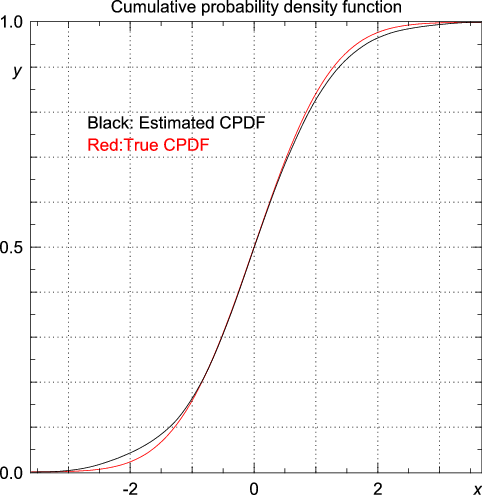
random module to generate 20000 random numbers from a distribution consisting of two Gaussians, one centered around -3 and one centered around 3, both with a standard deviation equal to unity. >>> from numpy.random import standard_normal, randint >>> n = 20000 >>> data = standard_normal(n) + 3.0*(randint(0,2,n)-0.5) >>> import statistics >>> y, x = statistics.pdf(data) 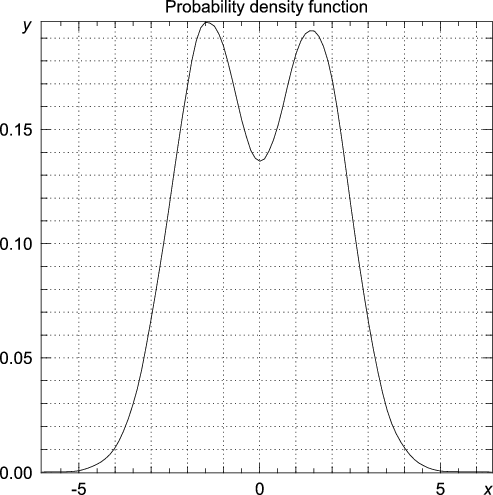
>>> y, x = statistics.pdf(data, kernel="Gaussian") 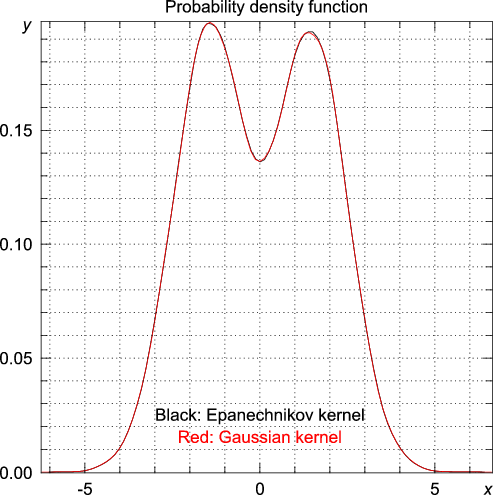
>>> statistics.bandwidth(data) 0.58133427540755089 >>> y, x = statistics.pdf(data, h = 0.58133427540755089/10) 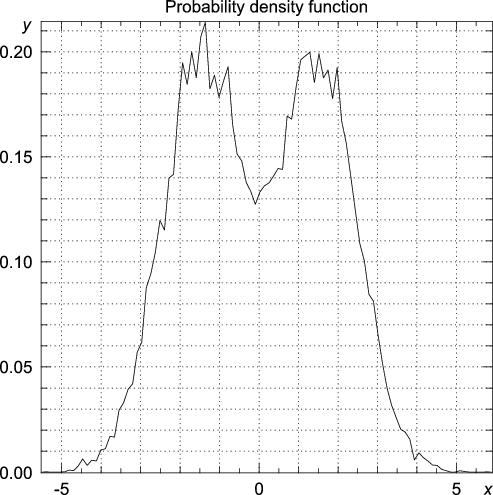
>>> y, x = statistics.pdf(data, h = 0.58133427540755089*4) 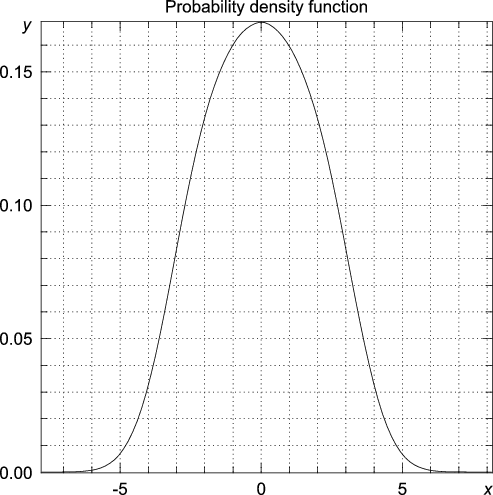
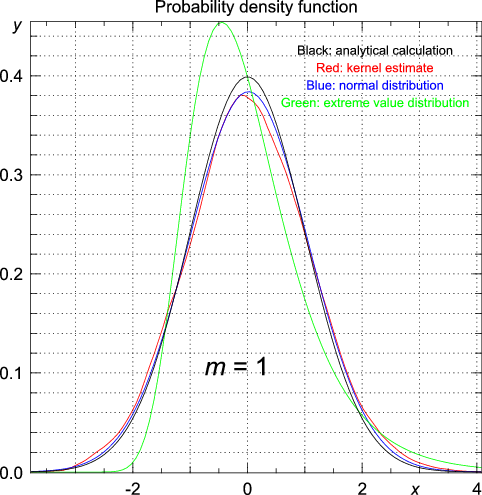
>>> import statistics >>> from numpy.random import standard_normal >>> n = 1000 >>> m = 1 >>> data = array([max(standard_normal(m)) for i in range(n)]) >>> y, x = statistics.pdf(data) 
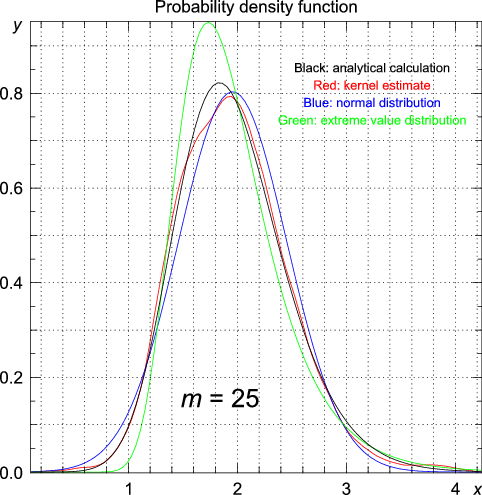
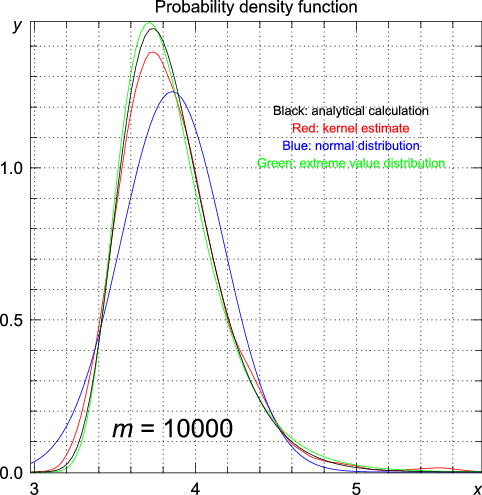
# using the data generated for m = 1 >>> statistics.cpdfc(data, x = 1.96) [ 0.02511014] | Exact (analytic) | Kernel estimate | Normal distribution | Extreme value distribution | |
| 1 | 0.0250 | 0.0251 | 0.0250 | 0.0497 |
| 3 | 0.0310 | 0.0310 | 0.0250 | 0.0465 |
| 5 | 0.0329 | 0.0332 | 0.0250 | 0.0411 |
| 10 | 0.0352 | 0.0355 | 0.0250 | 0.0451 |
| 25 | 0.0374 | 0.0377 | 0.0250 | 0.0409 |
| 100 | 0.0398 | 0.0396 | 0.0250 | 0.0514 |
| 200 | 0.0405 | 0.0407 | 0.0250 | 0.0482 |
| 1000 | 0.0415 | 0.0417 | 0.0250 | 0.0454 |
| 10000 | 0.0424 | 0.0427 | 0.0250 | 0.0506 |
>>> import numpy; print numpy.version.version gunzip statistics-<version>.tar.gz tar -xvf statistics-<version>.tar statistics-<version>. From this directory, type python setup.py config python setup.py build python setup.py install statistics-<version>.
For Python on Windows, a binary installer is available from http://bonsai.ims.u-tokyo.ac.jp/~mdehoon/software/python.